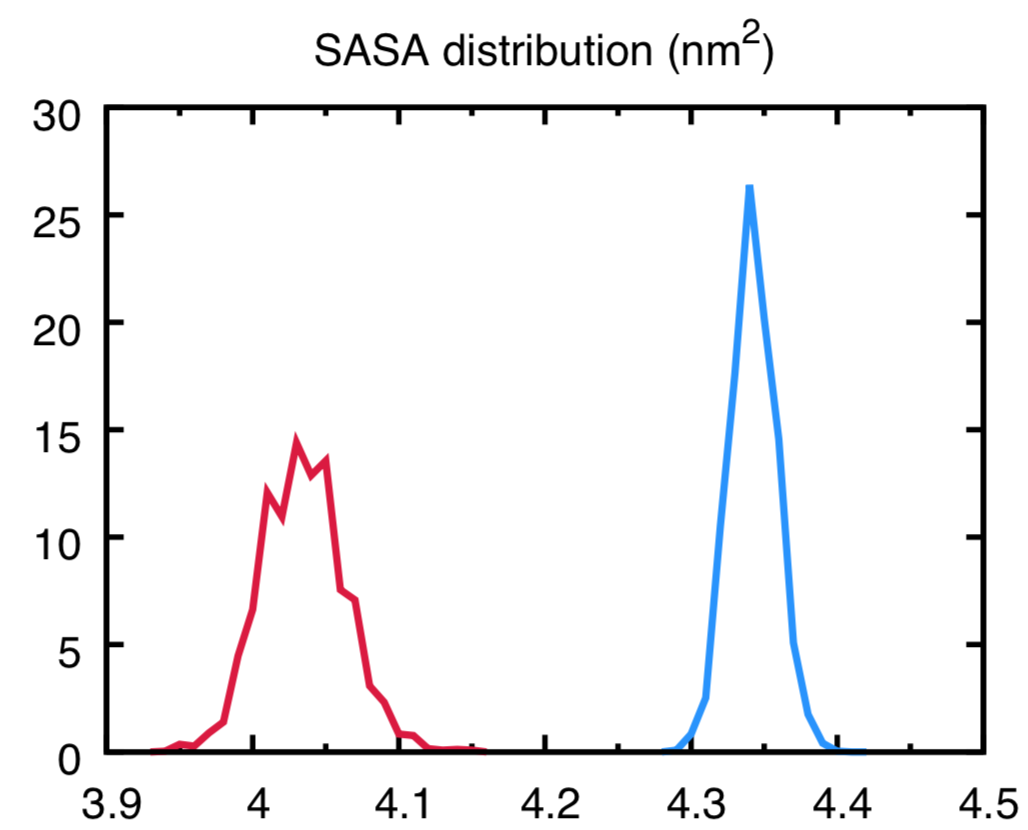
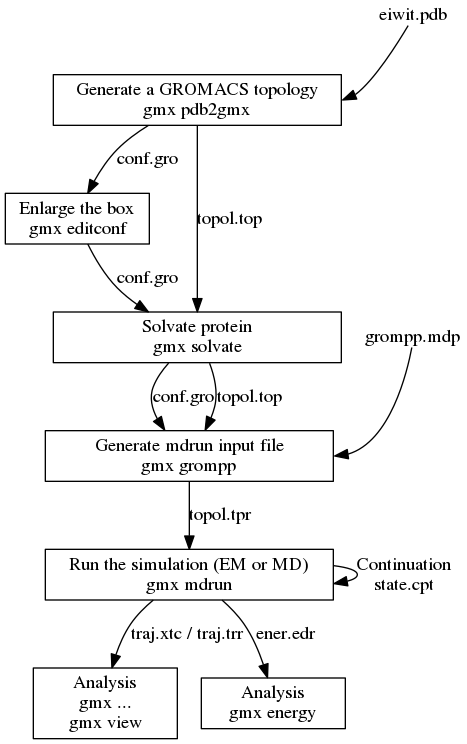
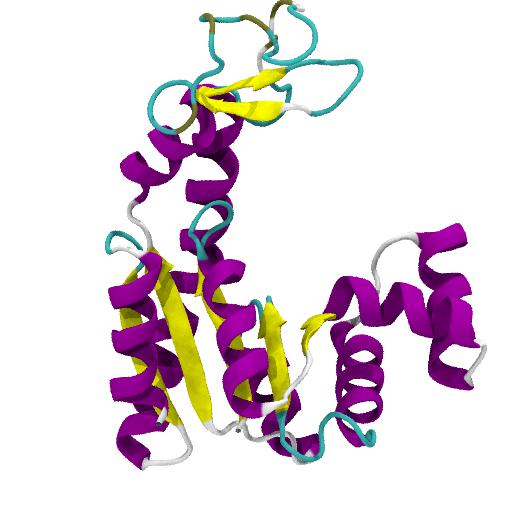
gmxapi: A GROMACS-native Python interface for molecular dynamics with ensemble and plugin support | PLOS Computational Biology
Intuitive, reproducible high-throughput molecular dynamics in Galaxy: a tutorial | Journal of Cheminformatics | Full Text